Research
Here you can find an overview of my current and past research projects.
Current Research
Hamiltonian Sparsity Testing (2025 - )
Keywords: hamiltonian testing, hamiltonian learning
Mentors: Professor Yu Tong (Duke University), Professor John Preskill (California Institute of Technology)
Description: A vital component of quantum device development is characterizing and calibrating its behavior. This can be accomplished by learning its Hamiltonian, taken to be a traceless, self-adjoint operator over \((\mathbb C^2)^{\otimes n}\), which can always be decomposed as follows:
\[\begin{equation*} H = \sum_{x \in \mathcal P_n} \lambda_x\sigma_x \end{equation*},\]where \(\mathcal P_n\) denotes the group of \(n\)-qubit Pauli operators.
While unconstrained learning is known to be computationally hard, it has been shown that imposing certain structural constraints, such as \(k\)-locality (meaning that each Pauli acts non-trivially on at most \(k\) qubits) or \(M\)-sparsity (meaning that the support of the Hamiltonian includes at most \(M\) Paulis), enable learning in polynomial time. Consequently, algorithms testing whether a given unknown Hamiltonian satisfies these properties are of utmost importance.
In this project, we aim to develop a sparsity testing algorithm that achieves Heisenberg-limited scaling, meaning that the total evolution time scales as \(\mathcal O(1/\epsilon)\).
🔗 Related Links (WIP):
- Senior Thesis
Phase Transitions in Projective Transverse Field Ising Models (2024 - )
Keywords: PTIM, phase transitions, entanglement
Mentor: Dr. Nat Tantivasadakarn (California Institute of Technology)
Description: Quantum error correction is an indispensable step towards scalable quantum computation. Errors and syndromes in quantum systems can be modeled as stochastic measurements, which act as non-commuting, competing projections. These measurements drive entanglement transitions between topological phases, delineating regimes where projective measurements either preserve or destroy quantum information. These transitions are commonly studied via the 1D projective transverse field Ising model (PTIM), a quantum variant of the classical Ising model wherein nearest-neighbor and external field interactions correspond to stochastic projective measurements of Pauli \(ZZ\) and \(X\) on a qubit chain. In this study, we extend the PTIM to \(\mathbb Z_4\) qudit chains featuring three non-commuting generalized Pauli operators to identify novel entanglement transitions.

Plot of half-chain entanglement entropy for different probabilities of measurement.

Plot of halfway mutual information for different probabilities of measurement.
🔗 Related Links (WIP):
- National Conference for Undergraduate Research 2025 Oral Presentation
- Southern California Conference for Undergraduate Research 2024 Oral Presentation
Past Research
Risk Factors for Metastatic Castration-Resistant Prostate Cancer (mCRPC) (2023 - 2024)
Keywords: mCRPC, cell-free DNA, androgen receptor
Mentor: Dr. Aadel Chaudhuri (Washington University in St. Louis)
Description: Prostate cancer is the second leading cause of cancer death among American men, causing 34,700 deaths annually. While localized prostate cancers are highly responsive to androgen-directed therapies, some patients develop metastatic castration-resistant prostate cancer (mCRPC), which is resistant to these treatments. Previous tumor whole-genome sequencing studies highlighted aberrations in both the androgen receptor (AR) locus and the recently discovered AR enhancer region as genomic hallmarks of mCRPC. The Chaudhuri Lab sought to replicate these results using cell-free DNA (cfDNA) analysis as a less invasive alternative to tumor sequencing, developing the EnhanceAR-Seq liquid biopsy assay to detect alterations in the AR locus/enhancer in post-treatment mCRPC plasma samples, which were found to portend primary resistance to treatment with high accuracy. In this study, we perform epigenomic analysis of pretreatment patient cfDNA samples, including genome-wide methylation sequencing, nucleosome profiling, and stemness analysis via EM-Seq, Griffin, and CytoTRACE, respectively, to elucidate the underlying biology of mCRPC. We illustrate that pretreatment plasma cfDNA analysis can be used to risk-stratify patients, mCRPC transcriptional profiles can be predicted from cfDNA epigenomics, and higher-risk mCRPC patients have more stem-like signature profiles that correlate with worse survival outcomes.
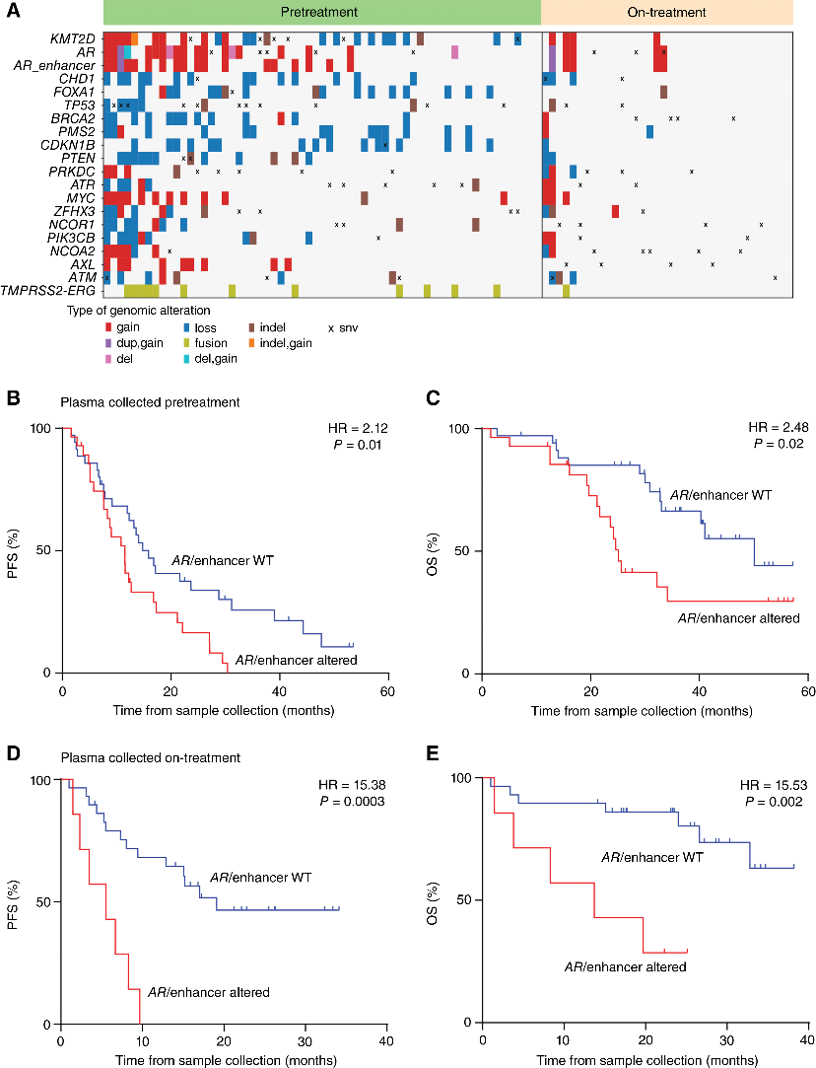
Genomic characterization of mCRPC plasma cfDNA. A, Genomic alterations detected in plasma cfDNA including the androgen receptor (AR) and the enhancer region upstream of AR in pre-ARSI and on-ARSI plasma collected from patients with mCRPC. PFS and OS Kaplan–Meier analysis according to AR/enhancer alteration status in plasma collected (B and C) before starting first-line ARSI treatment and (D and E) during first-line ARSI treatment. P values were calculated by the log-rank test, and HRs by the Mantel–Haenszel method. ARSI, androgen-receptor signaling inhibitor; cfDNA, cell-free DNA; mCRPC, metastatic castration-resistant prostate cancer; WT, wild-type. (figure from paper below)
🔗 Related Links:
- Published Paper
- Southern California Conference for Undergraduate Research 2023 Oral Presentation
- Summer Undergraduate Research Fellowship 2023 Oral Presentation
- WashU Radiation Oncology Symposium 2023 Oral Presentation
Evolution of Eukaryotic ATP Synthase (2020 - 2023)
Keywords: ATP synthase, eukaryotes, LECA
Mentor: Professor Jeremy Wideman (Arizona State University)
Description: Relatively little is known about ATP synthase structure in protists, and the investigated ones exhibit divergent structures distinct from yeast or animals. To clarify the subunit composition of ATP synthases across all eukaryotic lineages, we used homology detection techniques and molecular modeling tools to identify an ancestral set of 17 ATP synthase subunits. Most eukaryotes possess an ATP synthase comparable to those of animals and fungi, while some have undergone drastic divergence (e.g., ciliates, myzozoans, euglenozoans). Additionally, a ∼1 billion-year-old gene fusion between ATP synthase stator subunits was identified as a synapomorphy of the SAR (Stramenopila, Alveolata, Rhizaria) supergroup (stramenopile, alveolate, rhizaria). Our comparative approach highlights the persistence of ancestral subunits even amidst major structural changes. We conclude by urging that more ATP synthase structures (e.g., from jakobids, heteroloboseans, stramenopiles, rhizarians) are needed to provide a complete picture of the evolution of the structural diversity of this ancient and essential complex.
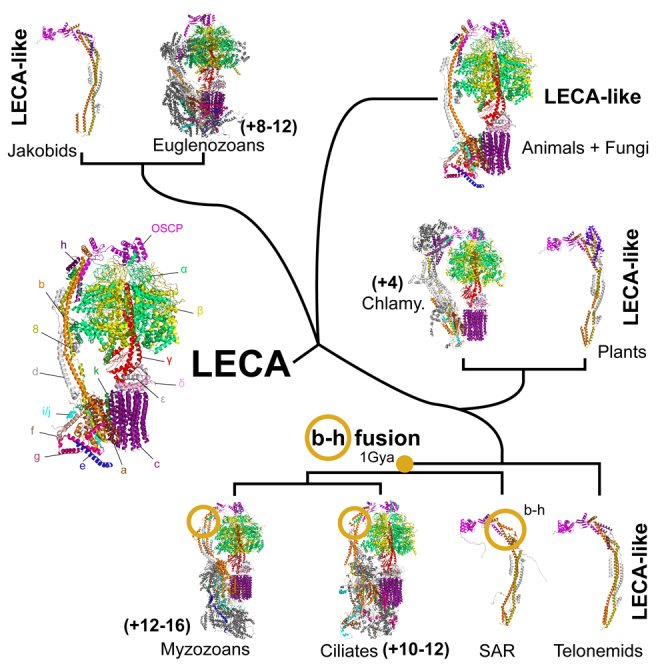
Diagram showing the reconstructed evolution of ATP synthase from the Last Eukaryotic Common Ancestor (LECA) (figure from paper below).
🔗 Related Links:
- First-Author Paper
- Additional Journal Publications:
- International Society of Protistologists 2021 Poster Presentation
- ASU Evolutionary Biology Symposium Oral Presentation